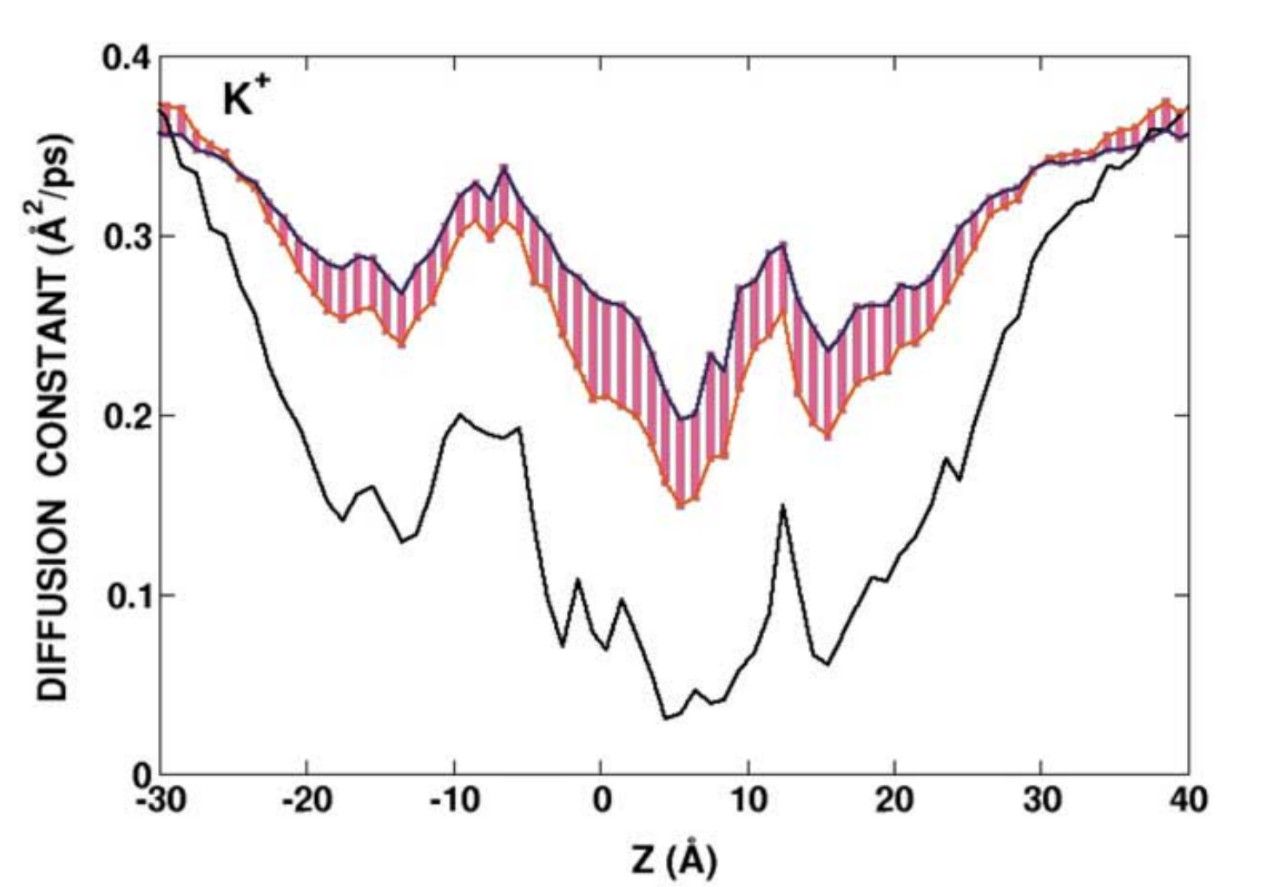
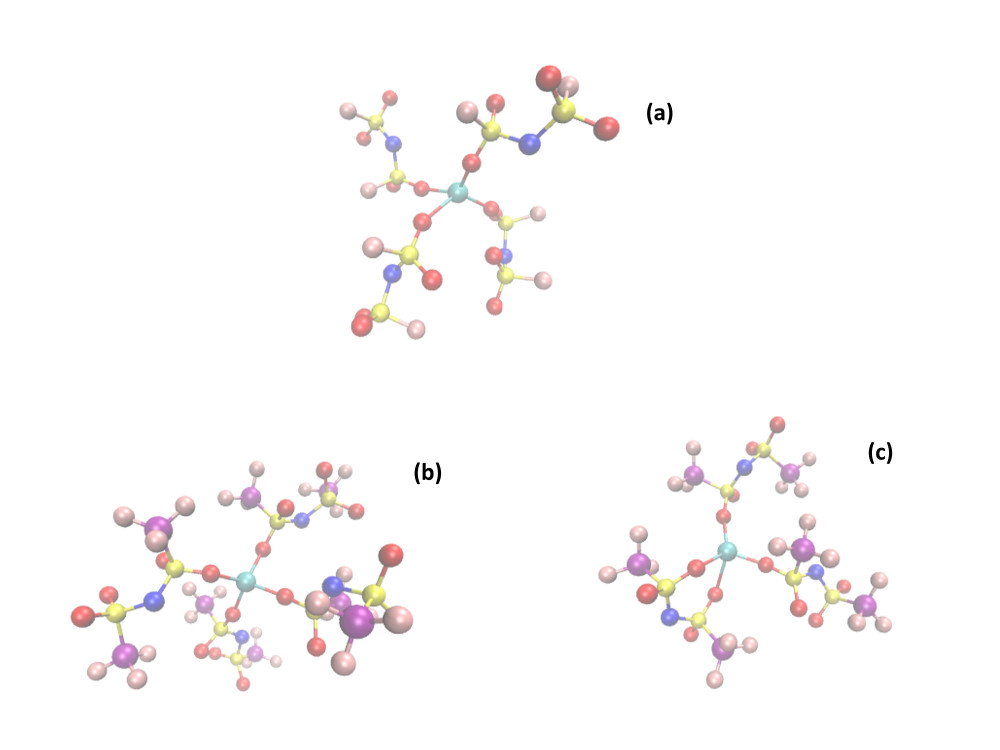
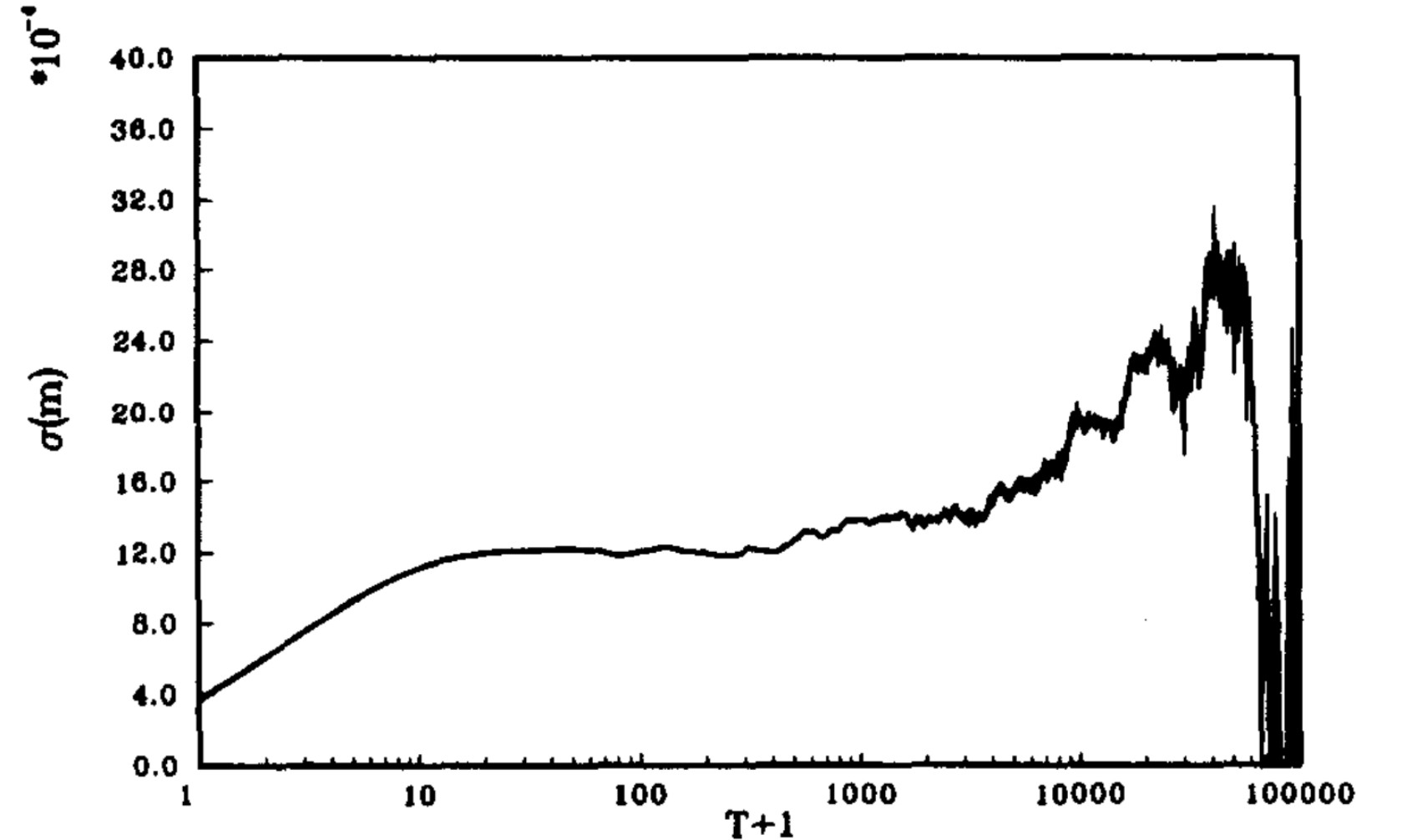
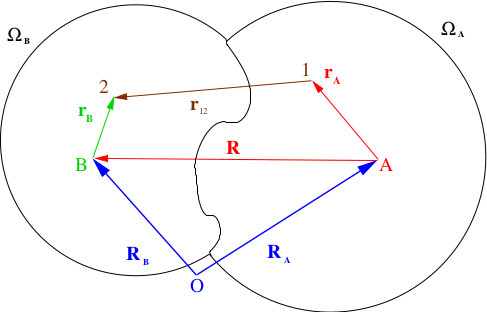
“Theoretical chemistry and Biophysics”
Over the years, I’ve gained broad experience in computational simulations and design of algorithms for studying organic (biological) and inorganic materials, which includes from methods based on quantum mechanics to methods based on classical and statistical mechanics. As a result, I’ve developed and implemented successful software solutions, some of them available in this website. In addition, some of my seminars, lectures and related material are also available.
“It seems to be one of the fundamental features of nature that fundamental physical laws are described in terms of a mathematical theory of great beauty and power (. . .)” P. A. M. Dirac
“The force of mind is only as great as its expression; its depth only as deep as its power to expand and lose itself.” G. W. F. Hegel
“The first principle is that you must not fool yourself – and you are the easiest person to fool.” R. P. Feynman
Name
Carlos José Fernández Solano
Address
Friederich-Humbert-Straße 159, Bremen (Germany)
Phone
+49 15118901525